Associate Research Professor
- B.Sc. Agra University, Agra, India
- M.Sc. Agra University, Agra, India
- Ph.D. Banaras Hindu University, Varanasi, India
- Postdoctoral Fellowship – Rutgers University

Building Address: 471b Bond Life Sciences Center
Phone Number: 573-882-9024
Email: singhka@missouri.edu
RESEARCH INTERESTS
The focus of my research is to study the basic science behind viral infections, mechanisms of susceptibility resistance of antivirals as well as drug-discovery against viruses and cancer. We study against immunodeficiency virus (HIV), hepatitis B and C viruses, Severe Acute Respiratory Syndrome Virus coronavirus (SARS-CoV), Middle East Respiratory Syndrome coronavirus (MERS-CoV), and (SARS-CoV-2) viruses. These pathogens are important and have claimed millions of lives around the world.
Our goal is to understand viral disease cycles using biochemical, structural, and virological tools. Thus, our efforts are directed to understanding the mechanistic details of viral components such as nucleic acid polymerases, helicases, and proteases. We employ fine biochemical tools such as steady- and presteady-state to decipher the kinetics mechanism of these critical viral enzymes. We investigate how resistance mutations affect the binding of approved drugs that lead the viruses to escape the impact of these drugs. Additionally, we use virological techniques to study the virus evolution under the pressure of antivirals. On the structural front, we use computational techniques to study the impact of mutations on the structures of viral proteins.
Another research project addresses the dynamics of the HIV virus around the world. We study the effectors of the varied efficacy of anti-HIV drugs on different subtypes. We study how does the efficacy of approved antiretrovirals (ARVs) is different in different HIV subtypes. We study the role of naturally occurring polymorphisms in the viral genome on the effect of ARVs. These studies are of high importance as they provide clinicians a guide to what drug should be prescribed to an HIV patient depending upon the subtype infection. We have successfully expressed and purified HBV polymerase (reverse transcriptase). The expression and purification of this polymerase has eluded scientists for several decades, and it has been a bottleneck in the drug-discovery against HBV. A complete characterization of HBV pol will most certainly expedite the drug-discovery against HBV.
In collaborations with Rutgers, we have discovered highly potent anti-cancer compounds that inhibit the binding of GAS6 protein to receptor tyrosine kinases. The interaction of GAS6 with RTKs (such as Tyro, Mer, and AXL) controls autophagy and contributes to many types of cancers.
TEACHING
- Virology 2020
- V-PBIO 5554 (VM-2s)
- V-PBIO 8454 (MPH)
SELECTED PUBLICATIONS (On NCBI)
- M. Pour, L. James, S. Mampunza, K. Singh*, F. Baer, J. A. Scott, M. G. Berg, M. A. Rodgers, G. A. Cloherty, J. Hackett Jr, and C. P. McArthur* (2020). Increased HIV in Greater Kinshasa Urban Health Zones: Democratic Republic of Congo (2017-2018). AIDS Res. Therapy (in press). *Corresponding authors.
- S. R. Kannan, A. N. Spratt, T. P. Quinn, X. Heng, C. L. Lorson, A. Sönnerborg, S. N. Byrareddy, K. Singh (2020). Infectivity of SARS-CoV-2: there Is Something More than D614G? J. Neuroimmune Pharmacol. (online doi: 10.1007/s11481-020-09954-3).
- S. Appelberg, S. Gupta, A. T. Ambikan, F. Mikealoff, A. Vegvery, S. Svensson-Akusjärvi, R. Benfeitas, M. Sperk, S. Krishnan, K. Singh ,J. M. Penninger, , A. Mirazimi, U. Neogi (2020) Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg. Microbes. Infect.9: 1748-1760.
- U. Neogi, K. J. Hill, A. T. Ambikan, X. Heng, T. P. Quinn, S. N. Byrareddy, A. Sönnerborg, S. G. Sarafianos and K. Singh. (2020). Feasibility of known RNA polymerase inhibitors as anti-SARS-CoV-2 drugs. Pathogens9: 320. (Cover Article)
- C. P. McArthur, F. Gallazzi, T. P. Quinn and K. Singh (2019). HIV capsid inhibitors beyond PF74. Diseases 7: 56
- A. Obasa, S. Mikasi, D. Brado, R. Cloete, K. Singh, U. Neogi, G. Jacobs (2019). Drug resistance mutations against protease, reverse transcriptase and integrase inhibitors in people living with HIV-1 (PLHIV) receiving second-line antiretroviral therapy on the South African National treatment program. Front. Microbiol. 11, 438.
- K. Singh* , F. Gallazzi, K. J. Hill, D. H. Burke, M. J. Lange, T. P. Quinn, U. Neogi and A. Sönnerborg (2019). GS-CA compounds: First-In-Class HIV-1 capsid inhibitors covering multiple grounds. Frontiers Microbiol. 10: 1227-1236. *Corresponding author.
- K. Singh* , S. G. Sarafianos, A. Sönnerborg (2019). Long-Acting anti-HIV drugs targeting HIV-1 reverse transcriptase and integrase. Pharmaceuticals, 12, 62. *Corresponding author. (Cover Article)
- A. D. Huber, D. L. Pineda, D. Liu, K. N. Boschert, A. T. Gres, J. J. Wolf, E. M. Coonrod, J. Tang, T. G. Laughlin, Q. Yang, M. N. Puray-Chavez, J. Ji, K. Singh, K. A. Kirby, Z. Wang and S. G. Sarafianos (2019). Novel hepatitis B virus capsid-targeting antiviral that aggregates core particles and inhibits nuclear entry of viral cores. ACS Infect. Dis. 5: 750-758.
- R. van Domselaar, D. T. Njenda, R. Rao R, A. Sönnerborg, K. Singh and U. Neogi (2019). HIV- 1 subtype C with PYxE insertion has enhanced binding of Gag-p6 to host cell protein ALIX and increased replication fitness. J. Virol. 93 : e00077-19.
- V. Shanbhag, K. Jasmer-McDonald, S. Zhu, A. L. Martin, N. Gudekar, A. Khan, E. Ladomersky, K. Singh, G. A. Weisman, M. J. Petris (2019). ATP7A delivers copper to the lysyl oxidase family of enzymes and promotes tumorigenesis and metastasis. Proc. Natl. Acad. Sci. USA. 116: 6836–6841.
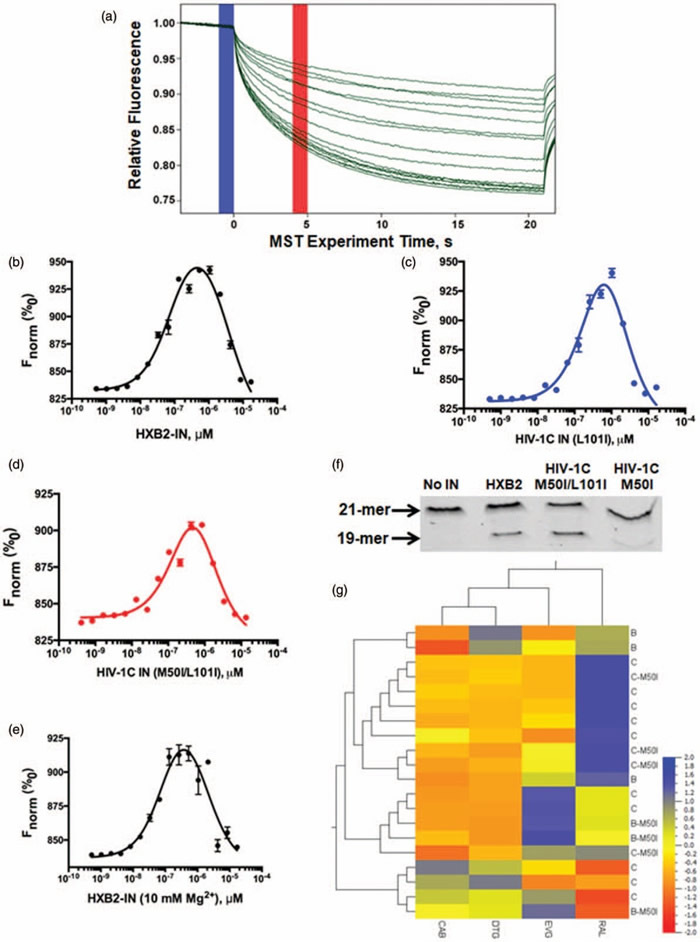
- K. J. Hill, L. C. Rogers, D. H. Burke, S. G. Sarafianos, A. Sönnerborg, U. Neogi, K. Singh . (2019). Strain-specific Effect on Biphasic DNA Binding by HIV-1 Integrase AIDS, 33: 588–592.
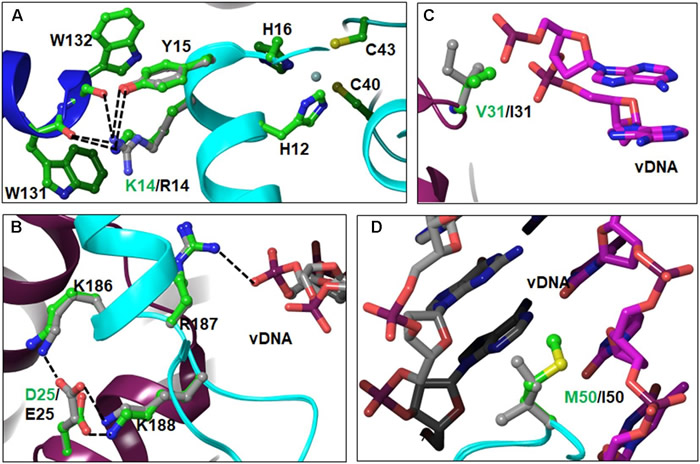
- L. Rogers, A. E. Obasa, G. B. Jacobs, A. Sönnerborg, U. Neogi, K. Singh. (2018). Structural implications of genotypic variations in HIV-1 integrase from diverse subtypes. Frontiers Microbiol. 9: 1954–1963.
- D. Brado, A. E. Obasa, G. M. Ikomey, R. Cloete, Singh K, S. Engelbrecht, U. Neogi and G. B. Jacobs (2018). Analyses of HIV-1 integrase sequences prior to South African national HIV- treatment program and available of integrase inhibitors in Cape Town, South Africa. Sci. Rep. 8:4709.